NovaSeq X User Guide: A Comprehensive Overview (Updated November 2, 2026)
This guide details the NovaSeq X Series, now featuring single-flow-cell systems and software v1.
New 25B kits boost multiomic workflows,
including single-cell sequencing.
The NovaSeq X Series represents a significant leap in high-throughput sequencing technology. Illumina’s innovative system, including the single-flow-cell NovaSeq X, dramatically increases data output and reduces costs per base. This series is designed for large-scale genomics projects, offering unparalleled speed and accuracy.
Recent updates, like software v1, further enhance the system’s capabilities. Novogene Europe’s investment in multiple NovaSeq X Plus systems demonstrates the growing demand for this technology. The introduction of new 25B kits expands multiomic applications, particularly in single-cell sequencing, making it a versatile platform for diverse research needs.
Understanding NovaSeq X System Components
The NovaSeq X system comprises several key components working in concert to deliver exceptional sequencing performance. Central to the system is the flow cell, available in various configurations, including the newly enhanced 25B format. These flow cells are paired with reagent kits, such as the 100-cycle and 200-cycle options, optimized for specific applications.
Illumina Experiment Manager (IEM) software controls the sequencing process, managing data acquisition and analysis. The system also includes fluidics and temperature control modules ensuring optimal reaction conditions. Proper understanding of each component is crucial for efficient operation and maximizing data quality.
NovaSeq X Plus: Enhanced Throughput and Capabilities
The NovaSeq X Plus significantly expands upon the already impressive capabilities of the NovaSeq X series, offering even greater throughput for large-scale genomic studies. Novogene Europe’s recent installation of a fifth NovaSeq X Plus system demonstrates the growing demand for this enhanced capacity.
This system excels in multiomic applications, particularly benefiting from the new 25B kits. These kits unlock enhanced capabilities for single-cell sequencing and metagenomics, allowing researchers to delve deeper into complex biological systems. The X Plus delivers faster turnaround times and reduced costs per sample.
Setting Up Your NovaSeq X System
Proper setup is crucial for optimal NovaSeq X performance. Initial installation and verification procedures must be followed meticulously to ensure system integrity. This includes confirming all components are present and functioning correctly, as per the manufacturer’s guidelines.
Network configuration is a key step, prioritizing data security throughout the process. Secure connections and appropriate access controls are essential for protecting sensitive genomic data. Careful planning and execution of these initial steps will lay the foundation for successful sequencing runs and reliable results.
Initial System Installation and Verification
Careful unpacking and placement are the first steps. Verify all components listed in the packing manifest are present and undamaged. Connect the system to a dedicated power source and ensure proper grounding. The initial boot-up sequence will initiate self-tests; monitor for any error messages.

Confirm network connectivity and access to Illumina support resources. Run the system verification suite provided by Illumina to validate instrument functionality. Document all installation details and verification results for future reference and troubleshooting. This meticulous approach ensures a stable foundation.
Network Configuration and Data Security
Secure network integration is paramount for NovaSeq X operation. Establish a dedicated network segment for the instrument, isolating it from public access. Configure static IP addresses and implement robust firewall rules to prevent unauthorized access. Enable data encryption both in transit and at rest, adhering to relevant data privacy regulations.
Implement strong user authentication protocols, including multi-factor authentication where possible. Regularly audit network logs for suspicious activity. Back up system configurations and sequencing data to secure, offsite locations. Prioritize data security to protect sensitive genomic information.
Workflow Overview: From Library Prep to Data Analysis
The NovaSeq X workflow encompasses several key stages, starting with meticulous library preparation. Following quality control, libraries are loaded onto flow cells for sequencing. Illumina Experiment Manager facilitates run setup and monitoring. Raw sequencing data undergoes base calling and demultiplexing, transforming it into usable reads.
Subsequent analysis involves variant calling and, potentially, genome assembly. Optimized pipelines are crucial for accurate results. This comprehensive process, from initial sample preparation to final data interpretation, ensures high-quality genomic insights.
Library Preparation for NovaSeq X
Effective library preparation is paramount for successful NovaSeq X sequencing. Recommended kits are designed to maximize compatibility and yield, ensuring optimal performance. Thorough quality control (QC) is essential; assess library size distribution and concentration before loading. Proper QC minimizes sequencing errors and maximizes data quality.
Utilizing validated protocols and reagents guarantees reliable results. Careful attention to detail during library construction directly impacts the accuracy and depth of sequencing data obtained from the NovaSeq X platform.
Recommended Library Prep Kits
Illumina offers a range of library prep kits optimized for NovaSeq X, enhancing multiomic capabilities. These include options for DNA and RNA sequencing, catering to diverse research needs. Specifically, kits supporting single-cell applications are now available, alongside those designed for metagenomics studies.
Selecting the appropriate kit depends on the starting material and desired application. Illumina’s documentation provides detailed guidance on kit selection and compatibility. Utilizing recommended kits ensures optimal performance and data quality on the NovaSeq X platform.
Quality Control of Libraries
Rigorous quality control (QC) is crucial before sequencing on NovaSeq X to ensure high-quality data. Recommended QC steps include assessing library size distribution using Bioanalyzer or TapeStation, and quantifying library concentration with Qubit or similar fluorometric methods.
Accurate quantification and size selection minimize sequencing errors and maximize data yield. Illumina provides detailed protocols for library QC. Addressing QC issues before sequencing saves time and resources, ultimately improving the reliability of downstream analyses.
Running a Sequencing Run on NovaSeq X
Initiating a sequencing run on the NovaSeq X requires meticulous preparation and adherence to established protocols. This begins with careful flow cell preparation and loading, ensuring proper fluidics and avoiding bubble formation. Subsequently, utilize Illumina Experiment Manager (IEM) to configure the run parameters.
IEM allows users to define read lengths, index read configurations, and output folder locations. Thoroughly review all settings before starting the run. Proper setup is paramount for optimal performance and data quality, maximizing the system’s throughput capabilities.

Flow Cell Preparation and Loading
Proper flow cell preparation is crucial for successful NovaSeq X runs. Begin by inspecting the flow cell for any visible damage. Gently remove the protective cap and ensure the flow cell surface is clean and free of debris. Carefully load the flow cell into the instrument, verifying secure placement.
Prime the flow cell according to the instrument’s prompts, ensuring complete wetting of the surface. Avoid introducing air bubbles during priming, as these can disrupt sequencing. Consistent and careful loading minimizes errors and maximizes data output.
Run Setup using Illumina Experiment Manager
Illumina Experiment Manager (IEM) v1 is central to NovaSeq X run configuration. Begin by creating a new experiment, specifying the flow cell type and read lengths. Define the sample sheet meticulously, including sample names, indices, and library concentrations. Ensure accurate sample information to avoid demultiplexing errors.
Review the run parameters, including output folder location and data format. Validate the experiment setup before initiating the run. IEM provides real-time monitoring capabilities, allowing for adjustments if needed. Proper setup guarantees optimal data generation.
Monitoring a Sequencing Run
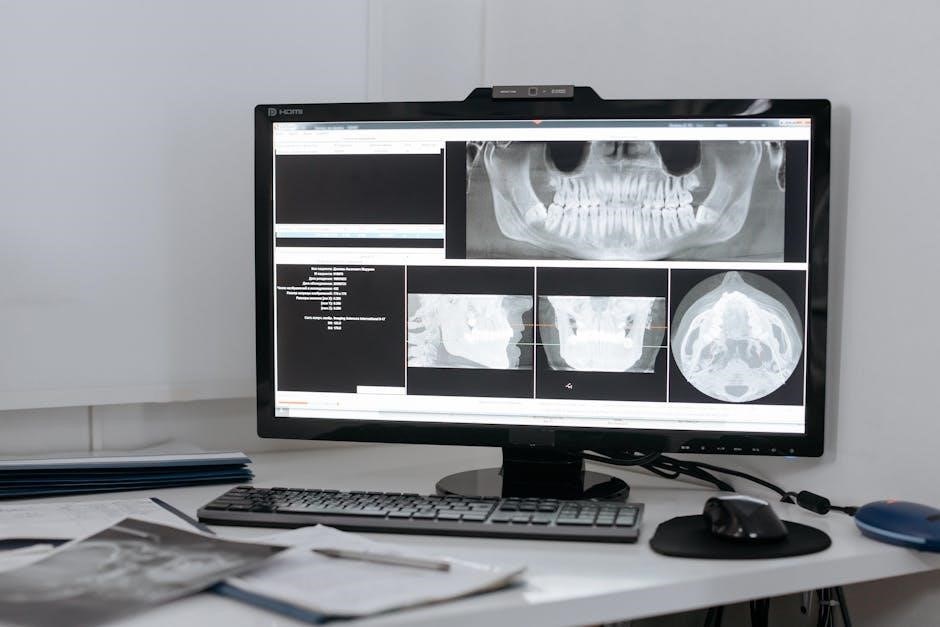
Real-time monitoring is crucial for NovaSeq X runs. Illumina Experiment Manager (IEM) displays key metrics like cluster density, error rates, and signal intensity. Regularly observe these parameters to identify potential issues early. Diagnostics tools within IEM assist in troubleshooting. Pay close attention to quality control metrics throughout the run.
Unexpected drops in signal or increases in error rates warrant immediate investigation. Utilize the troubleshooting resources available through Illumina support if problems arise. Proactive monitoring minimizes data loss and ensures high-quality sequencing results.
Real-Time Run Metrics and Diagnostics
Illumina Experiment Manager (IEM) provides comprehensive real-time data. Key metrics include cluster density, % passing filter, error rates (Q30 scores), and signal intensity over time. These are visualized graphically for easy interpretation. Diagnostics tools offer immediate assessment of potential issues, flagging anomalies like lane loading problems or reagent degradation.
Regularly review these metrics; deviations from expected values indicate potential problems. Utilize the built-in diagnostic reports for detailed analysis. Early detection allows for informed decisions, potentially saving valuable sequencing time and resources.
Troubleshooting Common Run Issues
Addressing issues promptly minimizes data loss. Common problems include low cluster counts (verify library concentration & flow cell quality), high error rates (check reagent freshness & instrument calibration), and lane dropouts (inspect flow cell for bubbles/damage). IEM provides error messages with suggested solutions.
If issues persist, consult the Illumina support website for detailed troubleshooting guides and knowledge base articles. Document all steps taken and error messages encountered before contacting Illumina support directly for assistance. Proactive documentation speeds resolution.
Data Analysis Pipelines for NovaSeq X
NovaSeq X generates substantial data requiring robust pipelines. Initial steps involve base calling, converting raw signal data into nucleotide sequences, and demultiplexing, separating reads by sample based on index sequences. Illumina’s bcl2fastq is commonly used for these processes.
Downstream analysis includes variant calling (identifying genetic differences) and genome assembly (reconstructing the complete genome). Specialized software like GATK or DRAGEN can be employed. Cloud-based solutions offer scalable compute resources for large datasets. Proper pipeline selection is crucial for accurate results.
Base Calling and Demultiplexing
Base calling transforms raw signal intensities into nucleotide sequences. Illumina’s software utilizes advanced algorithms to achieve high accuracy, crucial for downstream analyses. The process accounts for quality scores, indicating confidence in each base call. Demultiplexing then assigns reads to their original samples.
This relies on unique index sequences added during library preparation. Accurate demultiplexing is vital to avoid sample misassignment. Illumina Experiment Manager facilitates these steps, offering customizable parameters. Proper configuration ensures efficient and reliable data processing, setting the stage for meaningful insights.
Variant Calling and Genome Assembly
Variant calling identifies genetic differences—SNPs, indels—between a sample and a reference genome. Accurate variant identification requires robust algorithms and high-quality sequencing data, which NovaSeq X provides. Genome assembly reconstructs a complete genome from fragmented reads, a computationally intensive process.
Specialized software packages streamline these analyses, offering options for filtering, annotation, and prioritization of variants. The NovaSeq X’s high throughput accelerates both variant calling and de novo genome assembly projects. Careful parameter selection and validation are essential for reliable results.
NovaSeq X Software Updates and Management
Maintaining up-to-date software is crucial for optimal NovaSeq X performance and access to the latest features. Illumina Experiment Manager (IEM) v1 and subsequent versions provide a unified interface for run setup, monitoring, and data analysis. Regular software updates deliver improved algorithms, enhanced security, and compatibility with new kits.
Users should adhere to Illumina’s recommended software compatibility guidelines to avoid potential issues. Proactive software management ensures seamless operation and maximizes the value of your NovaSeq X investment. Check Illumina’s support website for the latest releases and documentation.
Illumina Experiment Manager (IEM) v1 and Beyond
Illumina Experiment Manager (IEM) v1 represents a significant advancement in NovaSeq X control and data handling. This software provides a streamlined workflow for designing and launching sequencing runs, monitoring progress in real-time, and initiating basic data analysis. Subsequent versions build upon this foundation, introducing enhanced features like improved sample sheet management and expanded reporting capabilities.
Users benefit from IEM’s intuitive interface and robust functionality, ensuring efficient operation of the NovaSeq X system. Staying current with IEM updates is vital for accessing the newest improvements and maintaining optimal performance.
Software Compatibility and Best Practices
Maintaining software compatibility is crucial for seamless NovaSeq X operation. Regularly check Illumina’s support website for the latest recommended software versions for both the NovaSeq X system and associated analysis tools. Prioritize using validated software combinations to avoid potential conflicts and ensure data integrity.
Best practices include establishing a robust data management system, implementing regular software backups, and adhering to Illumina’s guidelines for system updates. Proactive software management minimizes downtime and maximizes the reliability of your sequencing results.
NovaSeq X Kits and Consumables
The NovaSeq X system utilizes specialized kits and consumables for optimal performance. Currently available are NovaSeq X 25B kits, offered in both 100-cycle and 200-cycle configurations, designed to enhance multiomic applications. Proper flow cell storage is paramount; maintain them according to Illumina’s specifications to preserve reactivity and sequencing quality.

Always verify kit lot numbers and expiration dates before use. Utilizing genuine Illumina consumables ensures compatibility and reliable results. Careful handling of all components minimizes contamination risks and maximizes data output.
NovaSeq X 25B Kits (100-cycle and 200-cycle)
NovaSeq X 25B kits, available in 100-cycle and 200-cycle options, are optimized for high-throughput sequencing on the NovaSeq X platform. These kits significantly enhance multiomic capabilities, supporting a wide range of applications including whole-genome sequencing, RNA-Seq, and chromatin accessibility studies.
The choice between 100-cycle and 200-cycle kits depends on the read length requirements of your experiment. Longer read lengths are achieved with the 200-cycle kit, while the 100-cycle kit offers faster run times.
Flow Cell Storage and Handling
Proper flow cell storage and handling are crucial for optimal NovaSeq X performance and data quality. Flow cells should be stored at 2-8°C upon arrival and before use, ensuring they remain sealed to prevent contamination and dehydration. Avoid exposure to direct sunlight or extreme temperatures.
Before loading, inspect the flow cell for any visible damage. Once opened, flow cells must be used promptly. Refer to the detailed instructions within the NovaSeq X documentation for specific guidelines on flow cell preparation and loading procedures to maintain data integrity.
Multiomic Applications on NovaSeq X
The NovaSeq X platform excels in multiomic applications, offering enhanced capabilities for complex research. New 25B kits significantly improve workflows for simultaneous analysis of multiple data types, like DNA and RNA. This is particularly impactful for single-cell sequencing, enabling deeper insights into cellular heterogeneity.
Furthermore, NovaSeq X provides enhanced capabilities for metagenomics, allowing comprehensive profiling of microbial communities. These advancements facilitate a broader understanding of biological systems and accelerate discoveries across diverse research fields.
Single-Cell Sequencing with NovaSeq X
NovaSeq X dramatically advances single-cell sequencing, providing unprecedented throughput and sensitivity. The newly released Illumina Single-Cell Prep kits, coupled with the platform’s capacity, enable researchers to profile more cells than ever before. This allows for a more comprehensive understanding of cellular diversity and function.
The system’s high-resolution data generation facilitates the identification of rare cell populations and subtle differences between cells, crucial for disease research and drug discovery. NovaSeq X unlocks the potential for large-scale single-cell studies.
Enhanced Capabilities for Metagenomics
NovaSeq X significantly enhances metagenomic studies, enabling deeper and broader analyses of microbial communities. The platform’s high throughput allows for sequencing of complex environmental samples with greater coverage, revealing previously undetectable microbial diversity. This is vital for understanding ecosystem dynamics and identifying novel microbial functions.
Combined with new 25B kits, NovaSeq X facilitates comprehensive characterization of microbial genomes, improving our understanding of antibiotic resistance, host-microbe interactions, and environmental remediation strategies. Researchers can now explore metagenomes with unprecedented resolution.
NovaSeq X System Maintenance
Maintaining peak performance of your NovaSeq X system is crucial for reliable, high-quality sequencing data. Routine maintenance procedures, detailed in this guide, include regular cleaning of fluidics components and inspection of critical system parts. Adhering to the preventative maintenance schedule minimizes downtime and extends the lifespan of your investment.
Proper maintenance ensures optimal data output and reduces the risk of unexpected errors. Regularly scheduled checks and component replacements, as outlined, are essential for consistent performance. Following these guidelines will maximize the value of your NovaSeq X system.
Routine Maintenance Procedures
Daily checks involve inspecting fluid levels and verifying system status indicators. Weekly tasks include cleaning the flow cell chamber and inspecting tubing for any signs of wear or blockage. Monthly procedures encompass a thorough cleaning of the fluidics system using designated cleaning solutions, alongside filter replacements.
Regularly inspect and clean the optics to ensure optimal signal detection. Document all maintenance activities in the system log for traceability and troubleshooting. Following these procedures proactively prevents issues and maintains consistent sequencing performance, maximizing the lifespan of your NovaSeq X.
Preventative Maintenance Schedule
A tiered schedule ensures optimal NovaSeq X performance. Level 1 (monthly) includes fluidics system cleaning and filter changes. Level 2 (quarterly) involves a comprehensive inspection of all fluidic connections, pump functionality, and waste system integrity. Level 3 (annually) requires professional servicing, including optics alignment and a full system diagnostic check.
Adhering to this schedule minimizes downtime and extends system lifespan. Regularly updated software and firmware are crucial preventative measures. Detailed records of all maintenance are essential for tracking performance and identifying potential issues before they escalate, ensuring reliable sequencing results.
Troubleshooting Guide for NovaSeq X
This section addresses common issues encountered during NovaSeq X operation. Frequent error messages relate to flow cell communication or reagent delivery; consult the system logs for specific codes. Poor cluster registration can stem from library quality or improper flow cell loading. Unexpected data patterns often indicate reagent degradation or instrument malfunction.
For complex problems, detailed diagnostics are available through Illumina Experiment Manager. If issues persist, promptly contacting Illumina Support is recommended, providing comprehensive run information and error logs for efficient resolution and minimal downtime.
Common Error Messages and Solutions
Several error messages require immediate attention during NovaSeq X runs. “Flow Cell Error” often indicates a communication issue; reseating the flow cell or restarting the instrument may resolve it. “Reagent Cartridge Error” suggests a problem with cartridge installation or functionality – verify proper seating and sufficient reagent levels.
“Low Signal” can stem from library concentration issues or flow cell degradation. Consult the system logs for detailed error codes and corresponding troubleshooting steps within the Illumina documentation. If problems persist, contact Illumina Support with error details.
Contacting Illumina Support
Illumina provides comprehensive support resources for NovaSeq X users. For urgent issues impacting sequencing runs, initiate contact via the online support portal or dedicated phone lines, available 24/7. When reporting problems, provide detailed error messages, run parameters, and system logs for efficient diagnosis.
Access the Illumina website for extensive FAQs, knowledge base articles, and troubleshooting guides. Consider joining the Illumina Connect community forum to collaborate with peers and share solutions. Escalation paths are available for complex issues requiring specialized expertise.
Future Developments and NovaSeq X Roadmap
Illumina continues to innovate the NovaSeq X platform, focusing on enhanced throughput and expanded applications. Future software releases will prioritize improved data analysis pipelines and streamlined workflow integration. Expect advancements in multiomic capabilities, particularly for single-cell sequencing and metagenomics.
The roadmap includes exploring novel flow cell chemistries to further reduce costs and increase data yields. Illumina is committed to providing regular software updates and expanding the NovaSeq X ecosystem with new kits and consumables, ensuring long-term value for users.